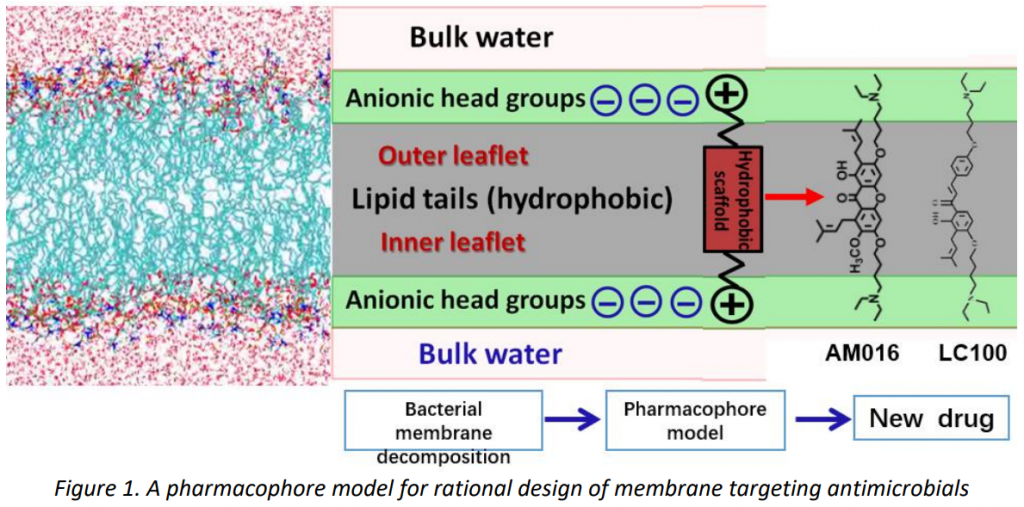
New analogues will be designed based on the atomistic insights from the simulations. The translation of in silico insights to practical antibiotic development will greatly accelerate the drug discovery process, providing cost saving measures, as well as the basis for a continued developmental process to meet new challenges presented by pathogens.
To find out more about how NSCC’s HPC resources can help you, please contact [email protected].
NSCC NewsBytes July 2023

New analogues will be designed based on the atomistic insights from the simulations. The translation of in silico insights to practical antibiotic development will greatly accelerate the drug discovery process, providing cost saving measures, as well as the basis for a continued developmental process to meet new challenges presented by pathogens.
To find out more about how NSCC’s HPC resources can help you, please contact [email protected].
NSCC NewsBytes July 2023